EQ 2x qPCR Master Mix Green is a reliable and robust real time PCR mix that is composed of a Hot Start DNA Polymerase, dNTPs, green-fluorescent dye and an optimized buffer system.
Please contact our exclusive distributor, Lab Supplies Scientific
EQ 2x qPCR Master Mix Green is a reliable and robust real time PCR mix that is composed of a Hot Start DNA Polymerase, dNTPs, green-fluorescent dye and an optimized buffer system. It has many applications in molecular analyses of DNA and RNA (cDNA) substrates, and it has been tested in commonly used real-time PCR instruments.
Tested for bacterial, human genomic DNA and human cDNA from HeLa cells, over at least 5 logs, of starting quantities of 1 ng (~ 2 X 10^5 copies), 64ng (~ 20 000 copies) and 40 ng respectively, the kit provides a high quantification efficiency (Efficiency >95%, and R2 >0.99) down to 1 copy of genomic DNA (4-6 copies of target genes) and 0.25 pg of cDNA templates (from Total RNA).
One of EQ 2x qPCR Master Mix Green’s most important features is its stability at 2-8 0C for at least 3 months after thawing, during which there is no observable loss of amplification efficiency.
Form: Liquid
Characteristics
Product Details
500 Reactions/ 20 μl
Reagents supplied:
|
RN014S |
||
| Cat No | Description | Tubes No. |
| RN013-1.3 | ΕQ 2x qPCR Μaster Μix Green, w/o ROX, 1.3 ml | 4 |
| BR033-1.8 | Nuclease Free Water, 1.8 ml | 2 |
Quality Control: The ΕQ 2x qPCR Μaster Mix Green kit, w/o ROXTM is functionally tested for its efficiency, as well as its absence of contaminating bacterial and human genomic DNA.
Storage: For long-term storage, store at -20 oC (up to 3 years). After thawing, store at 2-8oC for up to 3 months. Avoid multiple freeze-thaw cycles. Do not freeze-thaw more than 2 times.
Instrument compatibility:
Real-time instruments that do not require ROX™ internal reference dye as for example: Bio-Rad CFX OPUS TouchTM, CFX96 Touch™, CFX384 Touch™, CFX Connect™, DNA Engine Opticon® 2, Chromo4™, iCycler iQ™ and My iQ™, Roche LightCycler® 480, LightCycler® 1536, LightCycler® Nano, LightCycler® 96 and QuantStudio™ instruments, Thermo Scientific™ PikoReal™, Cepheid SmartCycler®, Bio Molecular Systems Mic qPCR cycler, Qiagen Rotor Gene Q, Rotor Gene 6000, MyGo Mini and MyGo Pro
Important Notes:
General tips for the Reactions’ setup may be found in the IFU (.pdf IFU)
Protocol
Standard Reaction Setup
Standard reaction may be found in Table 1.
| Component (Stock concentration) | Volume/Standard Reaction (proposed range) | Final Concentration in Standard Reaction (proposed range) |
| Nuclease Free Water | X μl | N/A |
| Primer A (10 μM) | 0.8 μl (0.2 -1.6 μl) | 0.4 μM (0.1-0.8 μΜ) |
| Primer B (10 μM) | 0.8 μl (0.2 -1.6 μl) | 0.4 μM (0.1-0.8 μΜ) |
| EQ 2x qPCR Master Mix Green, w/o ROX | 10 μl (you can also set 10 μl reaction by adding 5 μL and adjusting all other ingredient) | 1 X |
| Template DNA | Y μl (when high levels of PCR inhibitors are expected, do not use more than 10% of the reaction Volume)
|
Human Genomic DNA: 10 ng (100 ng-6 pg)
E. coli Genomic DNA: 1 ng (10ng – 10 fg) cDNA: 50 ng (100 ng-0.2 pg) Plasmid DNA: 0.5 ng (down to 3-5 copies) |
| Total Reaction Volume | 20 μl (or 10 μl) | N/A |
Table 1. Standard qPCR Reaction setup.
If using lower reaction volumes, scale all components proportionally. Reaction volumes < 10 µl are not recommended. Lower reaction volumes decrease signal intensity.
For each qPCR Reaction Master-mix, an additional 5% is recommended in the calculation. E.g., for 10 reactions, prepare a Master-mix for 10*1.05 =10.5 reactions. Round to the closest higher integer, 11 in the above example.
Always include positive and negative control reactions. A proposed order of template addition after aliquoting is to: a) add the negative control templates and close the caps, b) add the unknown sample templates and close the caps and finally c) add the positive control reactions’ templates.
qPCR Program
Three-step PCR Program
|
Two-step PCR Program
|
Table 2. Three-step and Two-step qPCR programs.
Initial Heat activation for 15 min is a very essential step!
REDUCING TIME OF INITIAL INCUBATION/ACTIVATION AT 95oC MAY AFFECT EFFICIENCY!
ΕQ 2x qPCR Μaster Μix Green kit, performance
ΕQ 2x qPCR Μaster Μix Green kit, w/o ROXTM has been repeatedly tested against several substrates:
Figure 1. Standard curve for E. coli DNA
Six, E. coli DNA 10-fold serial dilutions were prepared. 200 x 103 to 2 copies (1 copy of E. coli DNA weighs ~ 5.15 fg) were analysed in triplicates by RN014S kit and primers specific for 16S DNA. Images from CFX maestro 2.3 software of Bio-Rad. Left: Efficiency plot. Right: Amplification plot in logarithmic scale and melting curve plot.
Figure 2. Standard curve for Human Genomic DNA
Five, Human Genomic DNA 10-fold serial dilutions were prepared. 20 x 103 to 2 copies of haploid human genome (1 human haploid genome weighs ~3.2 pg) were analysed in triplicates by RN014S kit and primers specific for GAPDH genomic DNA. Images from CFX maestro 2.3 software of Bio-Rad. Left: Efficiency plot. Right: Amplification plot in logarithmic scale and melting curve plot.
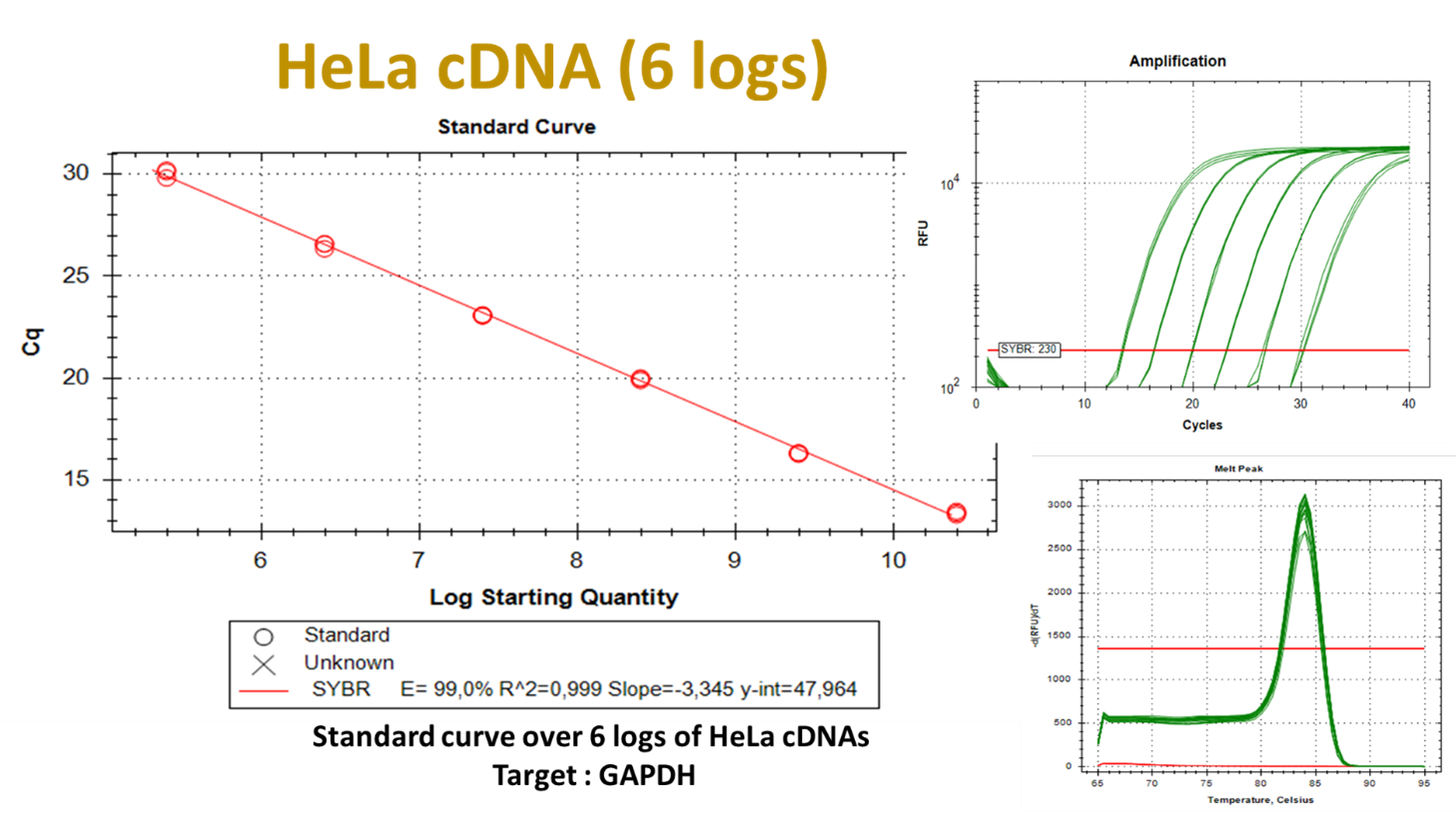
Figure 3. Standard curve for cDNA derived from HeLa total RNA.
Six, HeLa total cDNA, 10-fold serial dilutions were prepared. 25 ng/μl to 0.25 pg/μl were analysed by RN014S kit and primers specific for GAPDH, Exon7. Images from CFX maestro 2.3 software of Bio-Rad. Left: Efficiency plot. Right: Amplification plot in logarithmic scale and melting curve plot.
The above results (figures 1-3) have been produced in Bio-Rad qPCR instruments CFX96 and OPUS
EQ 2x qPCR Master Mix Green is a pre-mixed all-in-one 2x solution containing Hot Start DNA Polymerase, dNTPs, green-fluorescent dye, and an optimized buffer system. You just need to add primers and template - no need to prepare complex reaction mixtures.
One of the most important features is its stability at 2-8°C for at least 3 months after thawing, during which there is no observable loss of amplification efficiency. This eliminates the need for frequent freeze-thaw cycles.
Avoid multiple freeze-thaw cycles for optimal results. Do not freeze-thaw more than 2 times. After the initial thawing, store the remaining quantity at 4°C for up to 3 months. Quantification efficiency may drop by 5-8% after 2 freeze-thaw cycles.
This formulation (without ROX) is compatible with instruments that do not require ROX internal reference dye, including: Bio-Rad CFX series, Roche LightCycler series, Thermo Scientific PikoReal, Cepheid SmartCycler, Bio Molecular Systems Mic qPCR cycler, Qiagen Rotor Gene, and MyGo systems.
The optimal primer concentration range is 100-800 nM (0.1-0.8 μM). Start with 0.4 μM (0.8 μl of 10 μM stock in a 20 μl reaction). The optimal concentration is the lowest that produces the lowest Cq value and adequate fluorescence with minimal primer-dimer formation.
Initial heat activation at 95°C for 15 minutes is a very essential step. Reducing the time of initial incubation/activation at 95°C may significantly affect efficiency, especially for reactions with low template copies.